Chromatin Immunoprecipitation Sequencing (ChIP-seq) provides genome-wide profiling of DNA targets for histone modification, transcription factors, and other DNA-associated proteins. It combines the selectivity of chromatin immuno-precipitation (ChIP) to recover specific protein-DNA complexes, with the power of next-generation sequencing (NGS) to achieve high-throughput sequencing of the recovered DNA.
As protein-DNA complexes are recovered from living cells, binding sites can be compared in different cell types and tissues, or under different conditions. At Novogene, we offer high-quality sequencing and comprehensive bioinformatics solutions for your ChIP-seq projects.
Applications
ChIP-seq provides regulatory solutions and meets customized research needs, such as:
- Understanding how transcription factors regulate genes by profiling a genome-wide binding site and analyzing motif
- Outlining histone modification patterns associated with experimental treatment or sample conditions
- Investigating the relationships between epigenetic profiles and transcriptional regulations through joint analysis
- Accommodating to multiple immunoprecipitated DNAs using various methods, including ChIP, Cut&Tag, etc..
Benefits
- The output data is stable and reliable, with the guaranteed data quality of the sequencing score Q30≥ 80%.
- Expert bioinformatics analysis utilizes industry-standard MACS2 software and programs with the latest version to get motif prediction, peak annotation, functional analysis, and data visualization.
- Associated analysis is provided to explore correlations between ChIP-seq and gene expression.
Specifications: DNA Sample Requirements
| Sample Type | Required Amount | Fragment size | Purity |
| Enriched DNA Sample | ≥ 20 ng (Concentration ≥ 2 ng/μL) | 100 bp~500 bp | A260/280=1.8-2.0 |
Specifications: Sequencing & Analysis
| Platform | Illumina NovaSeq 6000 |
| Read Length | Pair-end 150 bp |
| Sequencing Depth | ≥ 20 million read pairs per sample for the species with reference genome |
| Standard Data Analysis |
|
Project Workflow
Novogene ChIP-seq service comprises four steps, including sample preparation, DNA library preparation, Illumina PE150 sequencing and data analysis using bioinformatics pipelines. From DNA sampling to obtaining data reports, each step may affect the quality and quantity of data output, directly affecting the results of subsequent bioinformatics analysis. Novogene strictly checks every step, including sample quality control, library quality control, and sequencing data quality control, to ensure the high quality, accuracy and reliability of sequencing data and provide comprehensive bioinformatics analysis.

Featured Publications using Novogene ChIP-seq Service
ChIP-seq provides genome-wide profiling of DNA targets for histone modification, transcription factors, and other DNA-associated proteins. Novogene provides high-quality sequencing and comprehensive bioinformatics analysis, including peak calling, motif analysis, peak annotation, differential analysis, and related functional analysis. We summarize a publication list with Novogene ChIP-seq service.
-
SET Domain–Containing Protein 4 Epigenetically Controls Breast Cancer Stem Cell Quiescence
Cancer ResearchIssue Date: 2019IF: 9.727DOI: 10.1158/0008-5472
-
Zscan4c activates endogenous retrovirus MERVL and cleavage embryo genes
Nucleic acids researchIssue Date: 15 July 2019IF: 11.501DOI: 10.1093/nar/gkz594
-
Nature communicationsIssue Date: 18 January 2019IF: 12.121DOI: 10.1038/s41467-018-08277-5
-
Cancer ResearchIssue Date: 2018IF: 9.727DOI: 10.1158/0008-5472
-
Targeting Epigenetic Crosstalk as a Therapeutic Strategy for EZH2-Aberrant Solid Tumors
cellIssue Date: 20 September 2018IF: 38.637DOI: 10.1016/j.cell.2018.08.058
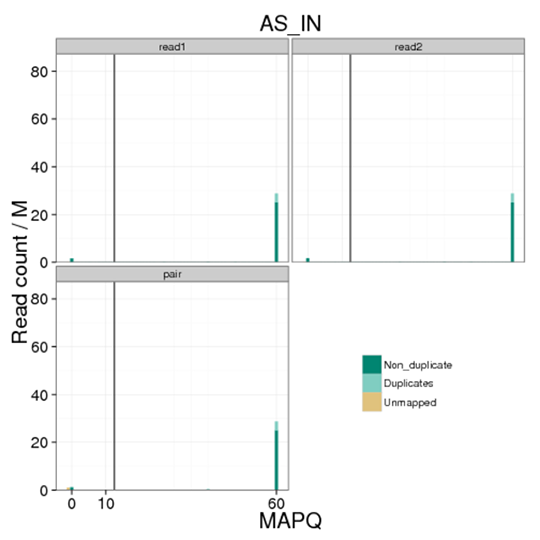
Distribution of MAPQ
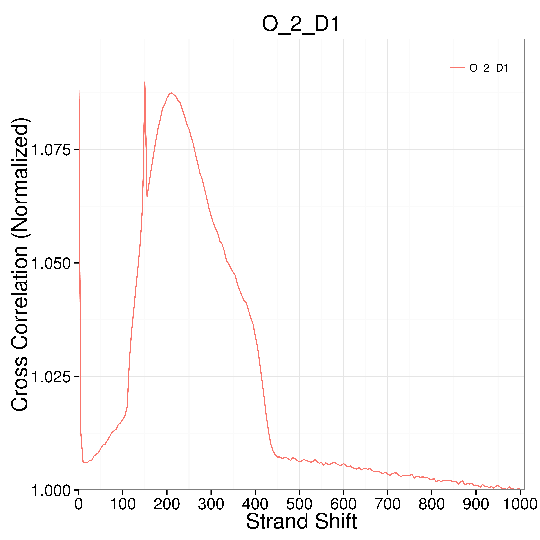
Plot of Strand Cross Correlation
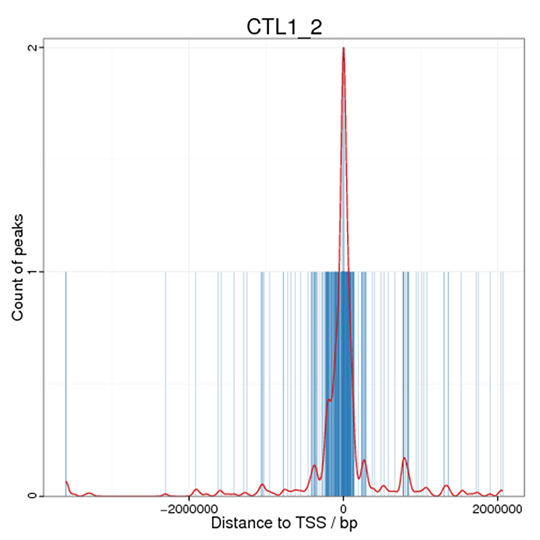
Distance Distribution of Peaks to TSS
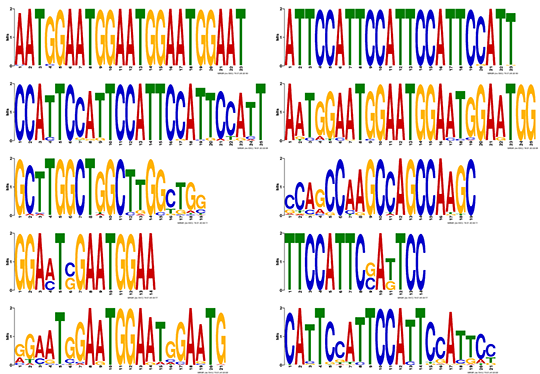
Motif Analysis
Peak Distribution in Functional Gene Regions
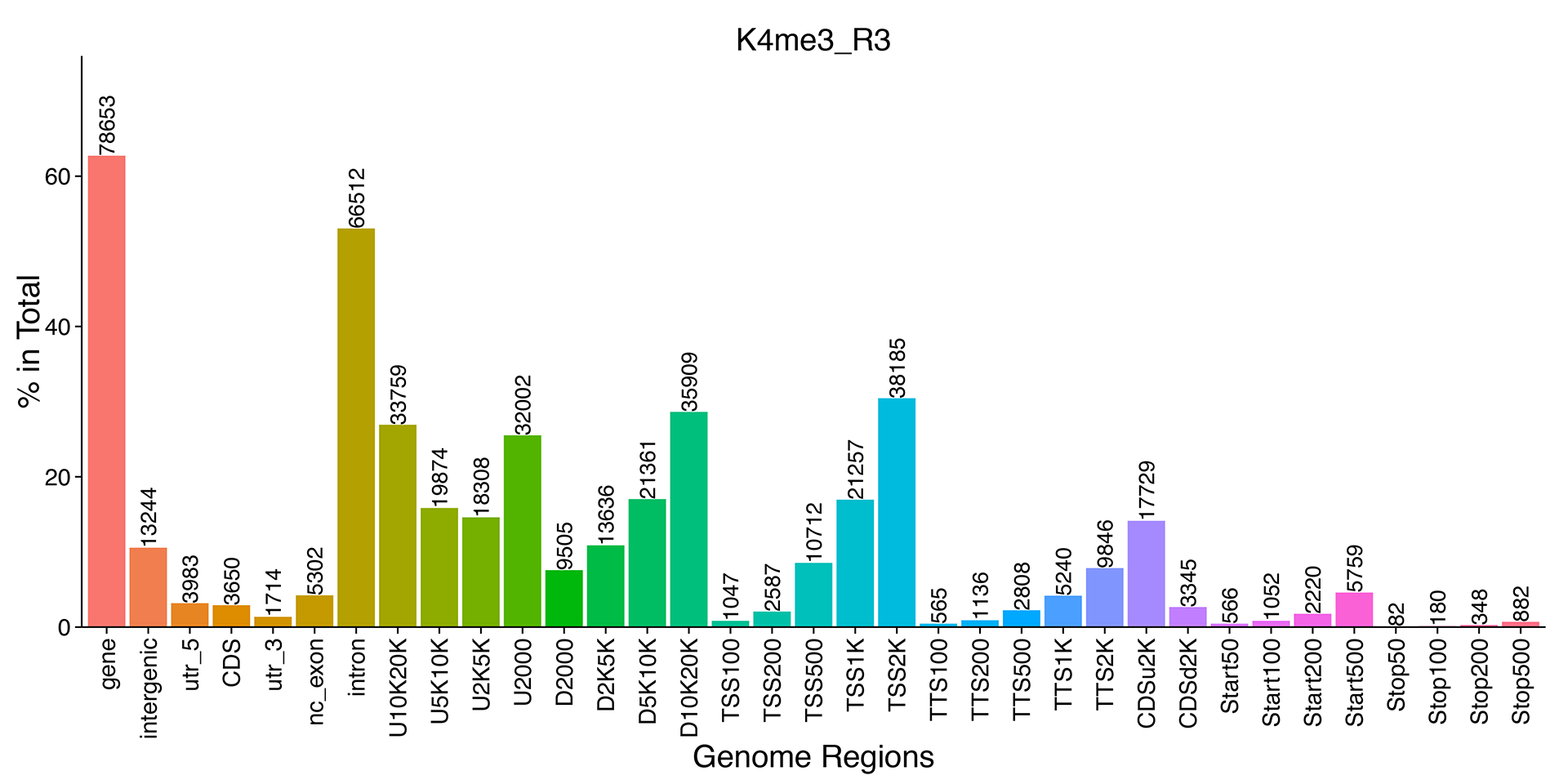
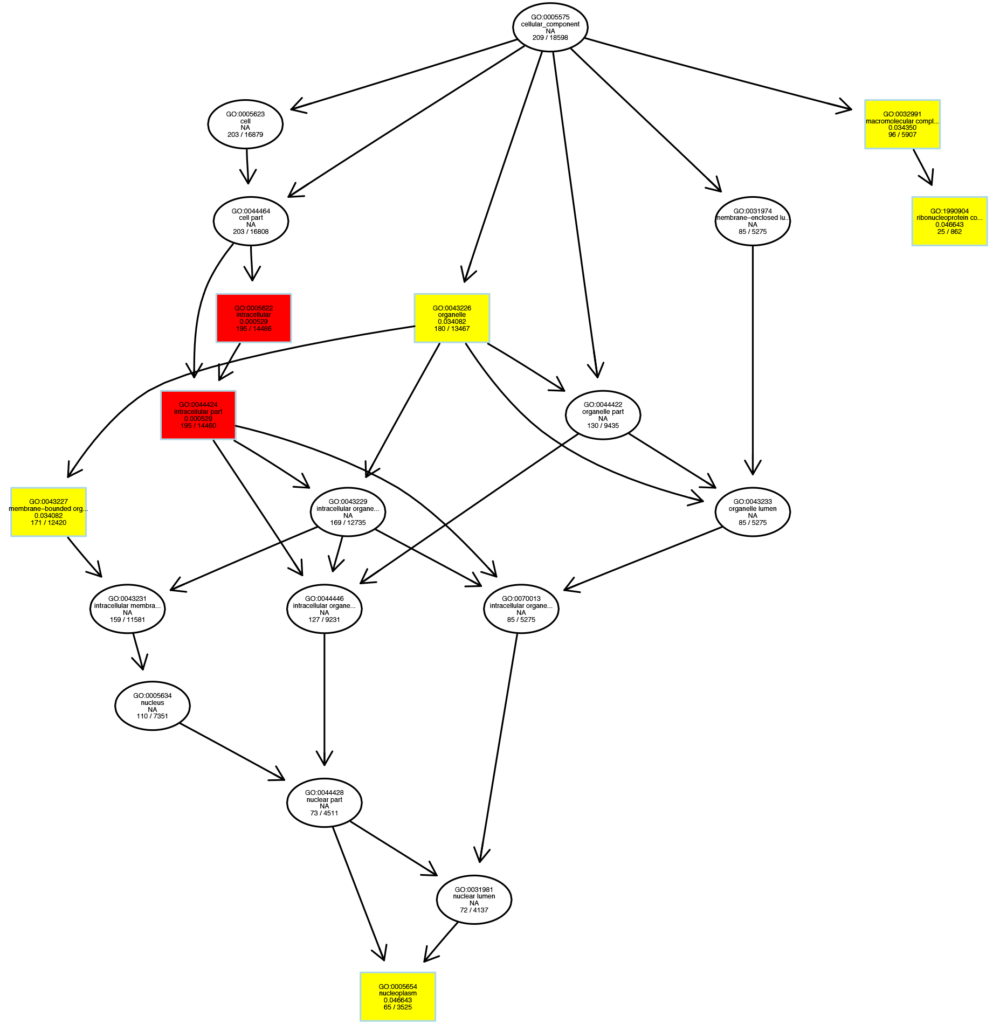
Diagram of GO Enrichment
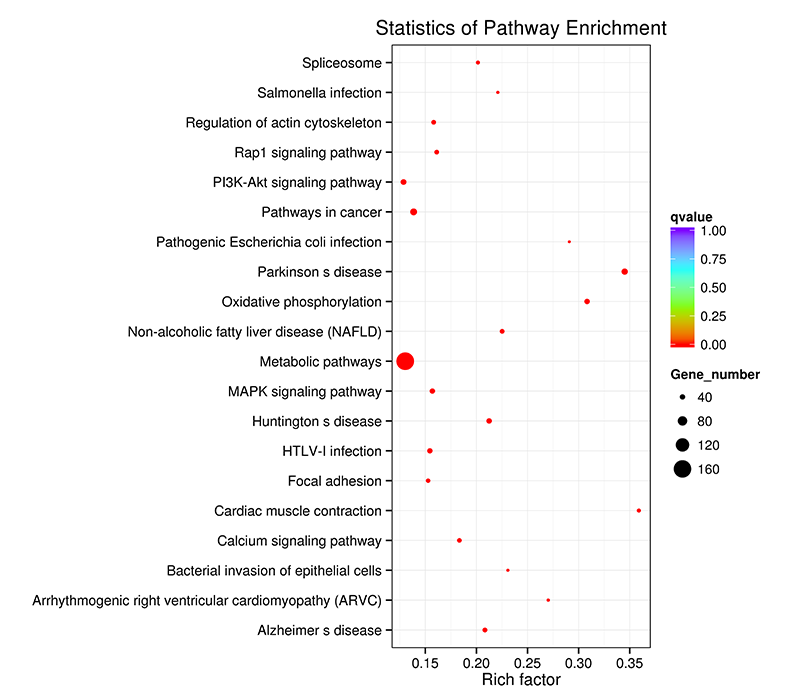
KEGG Enrichment Scatter Plot
More Services





