Proteins, essential to the fabric and function of the human body, are produced inside cells using genetic information contained in DNA. This article is the third in a four-part series on genes and chromosomes
Abstract
Proteins are made of chains of amino acids and form the largest organic component of the human body. They are essential not only to its fabric but also to its functioning. Instructions for building proteins are contained in the genetic code stored in deoxyribonucleic acid (DNA) in the nucleus of cells. To go from genes to proteins, a series of complex processes need to take place inside cells, including DNA transcription and translation. This third article in our series on genes and chromosomes examines how the genetic code stored in human genes is translated into proteins. It also explains how errors in the genetic code, or mutations, can lead to the production of abnormal proteins that may cause disease.
Citation: Knight J, Andrade M (2018) Genes and chromosomes 3: genes, proteins and mutations. Nursing Times [online]; 114: 9, 60-64.
Authors: John Knight and Maria Andrade are both senior lecturers in biomedical science at the College of Human Health and Science, Swansea University.
- This article has been double-blind peer reviewed
- Scroll down to read the article or download a print-friendly PDF here (if the PDF fails to fully download please try again using a different browser)
- Click here to see other articles in this series
Introduction
The blueprint for constructing and operating the human body is stored in sequences of deoxyribonucleic acid (DNA) called genes. This genetic information is used to make proteins with different functions, from structural proteins essential for building muscle, bone and skin to enzymes that catalyse the biochemical reactions essential to the body’s survival. Errors in the human genetic code, which are called mutations, sometimes lead to the production of abnormal proteins that may cause disease, including autoimmune disease and malignancy.
Proteins
Proteins are essential to both the fabric and function of the human body. These complex macromolecules are constructed from building blocks called amino acids, simple organic compounds containing a carboxyl (-COOH) and an amino (-NH2) group. There are 20 naturally occurring amino acids (Box 1); other variants can be produced synthetically in the laboratory.
Box 1. The 20 naturally occurring amino acids
Essential amino acids
- Histidine
- Isoleucine
- Leucine
- Lysine
- Methionine
- Phenylalanine
- Threonine
- Tryptophan
- Valine
Non-essential amino acids
- Alanine
- Arginine
- Asparagine
- Aspartic acid
- Cysteine
- Glutamine
- Glutamic acid
- Glycine
- Proline
- Serine
- Tyrosine
Amino acids
To function efficiently, human cells need to continually build new proteins to replace older, damaged ones. This requires a steady supply of all 20 amino acids. During digestion, the gastrointestinal tract sequentially breaks down the large animal and plant proteins contained in food into polypeptides, peptides and eventually into ‘free’ amino acids that are able to cross the gut wall to be absorbed into the bloodstream (VanPutte et al, 2017).
Naturally occurring amino acids (Box 1) are split into two categories:
- Essential – the nine amino acids that human cells canot synthesise and so need to obtain directly from food;
- Non-essential – the 11 amino acids that human cells can synthesise if direct supply through diet is low.
This terminology can be confusing, as so-called non-essential amino acids are actually essential for building proteins; the term is merely used to denote the fact that they do not have to be obtained from dietary intake.
If adequate nutrition is maintained through a healthy and balanced diet, cells receive the amino acids required for protein turnover. However, poor diet, eating disorders, certain medications and the ageing process (which can reduce appetite) can all restrict the dietary availability of amino acids, particularly the nine essential ones. This compromises the body’s ability to replace proteins, potentially resulting in muscle wastage and disease.
Protein varieties
Proteins form the largest organic component of the human body, making up approximately 50% of the dry mass of a typical human cell (Radivojac, 2013). They are synthesised in the cytoplasm of cells, where amino acids are linked together by peptide bonds to form long branching chains that range from a few amino acids to thousands. These amino acid chains further fold or twist into the unique three-dimensional configurations they need to adopt to fulfil their designated roles in the body (VanPutte et al, 2017).
The largest currently known protein in the human body is a muscle protein called titin (or connectin), which consists of around 33,000 amino acids. It works as a molecular spring and is thought to contribute to the force of muscle contraction (Powers et al, 2014).
The human genome project has shown that humans have just under 20,000 structural genes encoding for individual proteins. However, each gene can give rise to up to 100 variants of the protein it is coding for, so up to two million different varieties of proteins can be present in the human body (Ponomarenko et al, 2016).
Examples of common proteins include:
- Actin and myosin – contractile proteins found in muscle;
- Keratin – a dense protein found in hair, fingernails and the epidermis of the skin;
- Collagen – a general-purpose structural protein (used, for example, for building a framework of cartilage and bone) that can exist in various fibrous forms;
- Proteinaceous hormones such as insulin or glucagon – usually in short chains called peptides, these circulate in the blood and act as chemical messengers;
- Catabolic digestive enzymes such as pepsin, trypsin and amylase – these help digest and break up food macromolecules into simple components the body can absorb;
- Anabolic enzymes such as DNA polymerase, ribonucleic acid (RNA) polymerase and glycogen synthase – involved in the building of DNA, RNA and glycogen (animal starch) molecules;
- Haemoglobin – involved in the transport of oxygen and carbon dioxide in red blood cells (erythrocytes);
- Antibodies, often called immunoglobulins – small Y-shaped proteins that play a crucial role in the immune system, binding to foreign material, thereby labelling it for destruction;
- Neurotransmitters – small proteins that work as chemical messengers in the nervous system (for example, substance P);
- Neurotransmitter receptors – these (for example, the acetylcholine receptor) receive messages from neurotransmitters.
Ribosomes
Proteins are synthesised in the cells by ribosomes, small organelles densely arranged and embedded in the endoplasmic reticulum (ER) of the cytoplasm. The ER is a system of flattened interconnected membranes and most of its surface is covered by ribosomes, giving it a rough, uneven appearance that has led to the region being known as the rough ER. Here ribosomes ‘translate’ the genetic code by assembling amino acids to create proteins (Lewis, 2018).
Ribosomes are mostly composed of a specialised form of RNA called ribosomal RNA (rRNA), which is stabilised by small amounts of protein that also help assemble the ribosomes themselves (De la Cruz et al, 2015). In a single human cell, thousands of ribosomes can be actively constructing proteins at any one time.
From genes to proteins
Structural genes contain sequences of DNA determining the sequences of amino acid in proteins. Control (or regulatory) genes control which structural genes are expressed (‘switched on’) in a given tissue.
Using the genetic information contained in DNA to build proteins involves a few distinct steps:
- Transcription;
- Post-transcriptional modification;
- Translation;
- Post-translational modifications.
Transcription
The genetic information stored in the nucleus of a cell needs to be delivered to the ribosomes in the cytoplasm. Inside the nucleus of cells, DNA exists in the form of huge double helices. DNA molecules are too large to be passed on directly to the ribosomes so the genetic information stored in DNA needs to be copied onto a smaller, more mobile medium – that process is called transcription.
During transcription, a gene sequence coding for a protein is copied from double-stranded DNA onto single-stranded RNA (Fig 1). Single-stranded RNA molecules are much smaller than DNA molecules and can therefore travel through the tiny pores of the nuclear membrane. The role of RNA molecule will be to deliver protein-building instructions to a ribosome. As this form of RNA carries information across from the nucleus to the cytoplasm, it is called messenger RNA (mRNA).
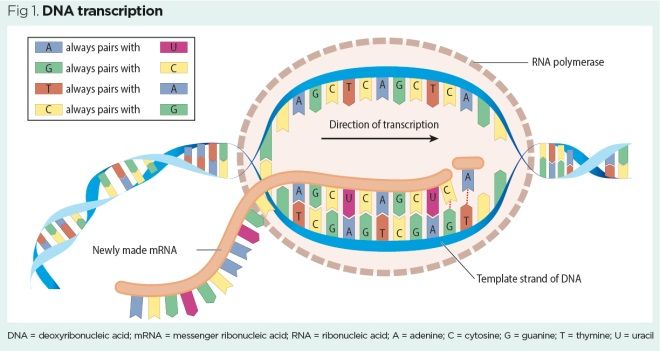
Like DNA, RNA is constructed from nucleotide bases, but unlike DNA it has no thymine base. In RNA, thymine is replaced by another nucleotide base called uracil. Base pairing rules in transcription differ slightly, therefore, from base pairing rules in DNA replication (Box 2).
Box 2. Complementary base pairing rules
In DNA replication (DNA to DNA)
- Adenine always pairs with thymine (A-T)
- Cytosine always pairs with guanine (C-G)
In DNA transcription (DNA to mRNA)
- Adenine always pairs with uracil (A-U)
- Guanine always pairs with cytosine (G-C)
- Thymine always pairs with adenine (T-A)
- Cytosine always pairs with guanine (C-G)
DNA = deoxyribonucleic acid. mRNA = messenger ribonucleic acid.
The process of DNA transcription is very similar to that of DNA replication (see part 2) and involves the following steps:
- The enzyme RNA polymerase binds to the beginning of the DNA gene sequence (also called promoter sequence);
- RNA polymerase unwinds a small portion of the DNA double helix to make it single-stranded (a process often described as analogous to undoing a zip) – the small area of unwound single-stranded DNA is called a transcription bubble (VanPutte et al, 2017);
- Only one of the exposed DNA strands holds useful information for constructing a protein – that strand is used as the template for transcription;
- RNA polymerase synthesises a complementary mRNA strand using the transcription base pairing rules.
Box 3 shows an example of transcription.
Post-transcriptional modifications
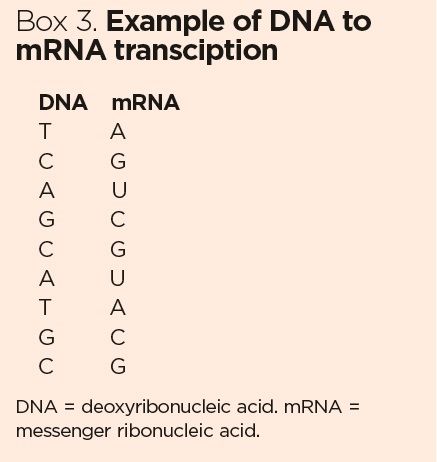
Not all information encoded in mRNA strands is useful for constructing a protein. A newly transcribed RNA strand consists of two elements:
- Exons – sequences of essential information for building a protein that has the correct sequence of amino acids;
- Introns – sequences that interrupt the exon coding sequences and do not usually hold useful information for building a protein. Once considered junk sequences, introns are now thought to play a role in regulating gene expression (Chorev and Carmel, 2012).
The introns need to be cut out and the exons spliced together to form a contiguous ‘high-fidelity’ mRNA sequence: this cutting and splicing – called post-transcriptional modification – is done by enzymes in the nucleus. The process is shown in Fig 2.
Nature of the genetic code
The genetic code is a triplet of three nitrogenous bases coding for one amino acid. As there are 20 naturally occurring amino acids, three bases allow for each amino acid to be represented by one triplet code (and some are represented more than once). The triplet code also allows ‘start-and-stop’ instructions to be encoded into the mRNA strand, so ribosomes know when to begin and when to end the construction of a protein.
Each run of three bases (triplet code) on an mRNA strand is called a codon. The first codon on any mRNA strand is always the ‘start’ codon – called AUG – which instructs the ribosome to begin protein synthesis. Since AUG also codes for the amino acid methionine, methionine is the first amino acid incorporated into a protein – if it is not actually needed, it will be removed later (Xiao et al, 2010).
The amino acids themselves are delivered to the ribosomes by transfer RNA (tRNA) molecules. A unique tRNA molecule corresponds to each amino acid and each tRNA molecule has its own unique triplet code, which corresponds to a codon on the mRNA strand. These tRNA sequences complement the mRNA codons and are therefore called anticodons (VanPutte et al, 2017).
Translation
After transcription and post-transcriptional modification, a mature, uninterrupted sequence of mRNA is generated. On entering the cytoplasm, this sequence attaches to a ribosome and can then be used for protein synthesis in a process called translation.
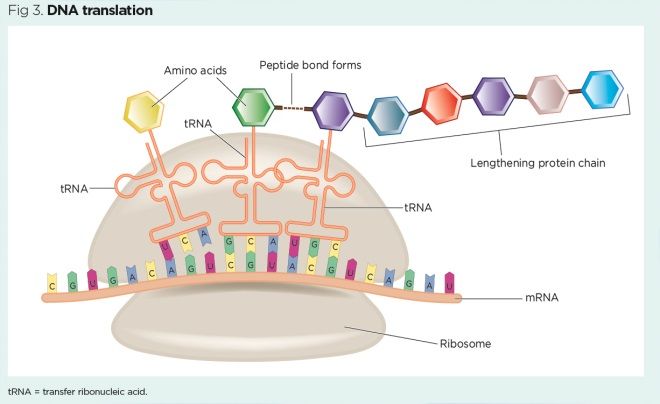
DNA translation (Fig 3) occurs a series of steps:
- A strand of mature mRNA attaches to a ribosome;
- Three bases on the mRNA strand (codon) are exposed on the ribosome – for example, AUG (the ‘start’ codon that also codes for methionine);
- A tRNA molecule arrives at the ribosome, aligns its anticodon to the codon on the mRNA strand and delivers the corresponding amino acid – for example, for the ‘start’ codon AUG, the corresponding anticodon UAC delivers methionine;
- The mRNA strand moves along the ribosome by three bases, exposing the next codon, then the next tRNA molecule arrives with its complementary anticodon and delivers another amino acid;
- Peptide bonds form between each adjacent amino acid and a protein chain starts to form;
- The mRNA strand continues to advance along the ribosome by three bases at a time, exposing each codon in turn. The tRNA molecules with corresponding anticodons continue to deliver amino acids, peptide bonds continue to form and the protein chain continues to lengthen;
- Eventually, a ‘stop’ codon (UAA, UAG or UGA) is reached at the end of the mRNA strand and protein synthesis is stopped – the process of DNA translation has created a crude protein.
Post-translational modifications
The crude protein usually needs to be modified before it can adopt its final 3D configuration and start performing its function in the body. These modifications occur in the cytoplasm in an organelle called the Golgi apparatus (also known as the Golgi body or simply the Golgi), which is often described as a cell’s ‘packaging and export area’.
Proteins undergo post-translational modifications as follows:
- The crude version of the protein is packaged into a small membrane-bound sac, the transfer vesicle;
- The transfer vesicle leaves the rough ER and migrates to the Golgi apparatus;
- In the Golgi apparatus, the crude protein is refined, which often involves adding sugar residues to the chain of amino acids via glycosylation (Huang and Wang, 2017) – many proteins in the human body actually are glycoproteins (proteins with added sugar);
- The refined protein leaves the Golgi apparatus and is either used in the cell or packaged into a secretory vesicle to be exported;
- Proteins designed to be exported are discharged from the cell as their secretory vesicle fuses with the plasma membrane.
Exported proteins can either be used in a tissue locally or transported to distant regions of the body by the blood. For example, the hormone insulin, synthesised in pancreatic beta cells, is released directly into the circulation when blood-glucose levels increase. It then functions as a chemical messenger binding to receptors (which are themselves proteins) on many human cells, instructing them to take up glucose, thereby normalising glucose concentration in the blood.
Mutations
To function correctly, proteins must have the correct sequences of amino acids, which ultimately relies on the genetic code remaining constant. However, there are so many nucleotide bases in the human genome (approximately three billion base pairs) that errors invariably occur. Such errors are referred to as mutations and can lead to the production of proteins that may not function correctly. Abnormal proteins are associated with a variety of diseases, including some forms of autoimmune disease and malignancy.
Genetic mutations may occur randomly following errors in DNA replication (described in part 1 of this series), particularly as the body ages; alternatively, they may be caused by environmental factors that directly damage DNA molecules. Anything that can damage DNA and lead to mutation is called a mutagen (VanPutte et al, 2017). Many genetic mutations occur in sections of DNA that do not code for proteins (for example, in the non-coding introns), so they usually have little impact on physiological function.
Factors known to damage DNA, and therefore increase the risk of mutation, include:
- Increasing age;
- Pollutants;
- Infections – particularly viral infections, as viruses often insert their genes into human DNA, potentially interrupting gene sequences;
- Radiations – for example, ultraviolet (UV) light from the sun or X-rays from medical imaging.
UV light from the sun (particularly UVB) is known to damage DNA in skin cells. If mutation occurs in the control genes that regulate cell division or in the genes that code for DNA repair enzymes, the result can be uncontrolled cell division and skin cancer (Hopkins, 2015). Although human skin exposed to sunlight produces its own natural UV protection in the form of melanin (the dark pigment that tans the skin), depletion of the ozone layer and excessive time in the sun can lead to damaging doses of UV radiation that increase the risk of skin cancer. Sunscreens offer better UV protection and have been shown to significantly reduce UV-induced skin damage and skin cancers (Green and Williams, 2007).
Mutations such as those caused by UV radiation to DNA in the skin are not generally passed down through generations. However, when mutations affect the germinal cells of the testes and ovaries, they can be inherited by offspring. Over 100,000 mutations have been reported in human germinal cells, many of which are associated with common heritable genetic diseases (Vipond, 2013; Cooper et al, 2010). Some of these will be explored in the fourth and final article in this series.
Key points
- Deoxyribonucleic acid (DNA) stored in genes contains the blueprint for building the human body
- Proteins, essential to the fabric and function of the body, are chains of amino acids
- Proteins are synthesised in cells by the ribosomes
- Protein synthesis involves DNA transcription, post-transcriptional modification, translation and post-translational modifications
- Genetic mutations can lead to abnormal proteins, which in turn can lead to autoimmune disease and malignancy
Also in this series
Cooper DN et al (2010) Genes, mutations, and human inherited disease at the dawn of the age of personalized genomics. Human Mutation; 31: 6, 631-655.
De la Cruz J et al (2015) Functions of ribosomal proteins in assembly of eukaryotic ribosomes in vivo. Annual Review of Biochemistry; 84: 93-129.
Green AC, Williams GM (2007) Point: sunscreen use is a safe and effective approach to skin cancer prevention. Cancer Epidemiology, Biomarkers and Prevention; 16: 10, 1921-1922.
Hopkins R (2015) How Ultraviolet Light Reacts in Cells. SciBytes.
Huang S, Wang Y (2017) Golgi structure formation, function, and post-translational modifications in mammalian cells. F1000 Research; 6: 2050.
Lewis R (2018) Human Genetics: Concepts and Applications. New York, NY: McGraw-Hill Education.
Ponomarenko EA et al (2016) The size of the human proteome: the width and depth. International Journal of Analytical Chemistry; doi: 10.1155/2016/7436849.
Powers K et al (2014) Titin force is enhanced in actively stretched skeletal muscle. Journal of Experimental Biology; 217: 3629-3636.
Radivojac P (2013) A (Not So) Quick Introduction to Protein Function Prediction.
VanPutte CL et al (2017) Seeley’s Anatomy and Physiology. New York, NY: McGraw-Hill Education.
Vipond K (2013) Genetics: A Guide for Students and Practitioners of Nursing and Health Care. Banbury: Lantern Publishing.
Xiao Q et al (2010) Protein N-terminal processing: substrate specificity of Escherichia coli and human methionine aminopeptidases. Biochemistry; 49: 26, 5588-5599.


Have your say
or a new account to join the discussion.