1. Introduction
The Japanese indigenous species Angelica acutiloba Kitagawa var. acutiloba Kitagawa (Toki) or A. acutiloba Kitagawa var. sugiyamae Hikino (Hokkai Toki) are listed in the Japanese Pharmacopoeia (JP), 16th edition [1], and are important medicinal materials for traditional Japanese medicine. The cultivar A. acutiloba var. acutiloba, which is generally considered to be higher quality than the “Hokkai Toki”, was originally grown in Nara Prefecture, Japan. The difference in the quality of the two cultivars has been described in two Japanese historical manuscripts [2,3]. To this day, the quality of A. acutiloba var. acutiloba is still regarded as being higher than that of other cultivars, and this is reflected in the market price of the root. Since A. acutiloba var. sugiyamae “Hokkai Toki” has been cultivated in various regions of Japan including Nara Prefecture since the 1950s [4], several Angelica strains are thought to be outcrossed [5]. Furthermore, A. acutiloba Kitagawa var. iwatensis Hikino (Miyama Toki), which is genetically very close to ‘Toki’, and A. stenoloba
(“Hosoba Toki” or “Tokachi Toki”) grow wild in Japan [6]. Because these species and varieties are closely related to A. acutiloba, they readily outcross with A. acutiloba var. acutiloba and A. acutiloba var. sugiyamae [7]. The root of A. acutiloba var. iwatensis is now not used as a crude drug listed in JP 16th edition, although the morphological features of the roots are very similar among related species of A. acutiloba var. acutiloba and it is difficult to reliably distinguish them from each other. A previous molecular survey of chloroplast DNA and the internal transcribed spacer of nuclear ribosomal DNA showed some nucleotide polymorphisms that could be used to distinguish between A. acutiloba var. acutiloba and A. acutiloba var. sugiyamae [8]. Although genomic DNA polymorphisms revealed differences among Angelica varieties, an efficient and simple discrimination method that can identify varieties of A. acutiloba is needed.
The random amplified polymorphic DNA (RAPD) method can be used to study the genetic variability of species, subspecies, varieties, and natural populations [9-12]. The advantage of RAPD analysis is that it is simple, fast [13], and it does not require genomic information [14]. A previous RAPD analysis showed some polymorphism for discriminating A. acutiloba var. acutiloba and A. acutiloba var. sugiyamae, but the polymerphism of A. acutiloba var. sugiyamae were quite similar to those of A. stenoloba [15]. Therefore, clear discrimination marker for A. acutiloba varieties and related species was needed. In this study, we report the authentication and genetic variation among A. acutiloba varieties and related species as determined using RAPD markers.
2. Materials and Methods
2.1. Plant Materials
The plant materials included nine strains of Angelica acutiloba var. acutiloba, three of A. acutiloba var. sugiyamae, and two of A. acutiloba var. iwatensis from cultivars grown in different locations or native populations. Cultivated strains of the related species A. stenoloba (“Hosoba Toki”), A. stenoloba f. lanceolata (“Tokachi Toki”), and A. shikokiana (“Inu Toki”) were also collected from various locations in Japan (Table 1). All strains/varieties were cultivated in a greenhouse before use in experiments. Each collected plant was identified by its morphological characters.
2.2. Total DNA Extraction
Total DNA was extracted from young leaf tissue (approx. 200 mg) using a modified CTAB method [16]. The DNA concentration was determined by absorbance at 260 nm.
2.3. RAPD Analysis
We used 80 decamer (10-mer) primers (OPA, OPB, OPC, and OPD; Operon Technologies, USA, Table 2) in the RAPD analysis. Each 30-μ l PCR mixture contained 200 ng DNA, 1 × ExTaq buffer, 0.2 mM each deoxynucleotide triphosphate, 1 mM each primer, and 2.5 U ExTaq (TaKaRa, Japan). Amplification was performed using a PTC-100 Programmable Thermal Controller (MJ Research Inc.), as follows: initial denaturation for 5 min at 95˚C, followed by 42 cycles of 95˚C for 30 s, 40˚C for 1 min, and 72˚C for 2 min, and a final extension at 72˚C for 10 min. RAPD fragments were separated electrophoretically in 1% agarose gels, stained with ethidium bromide, and visualized on a UV transilluminator. To optimize the RAPD analysis, we trialed PCR mixtures containing 20, 40, 120, 200, or 400 ng total DNA, and 1, 2.5, or 5 units ExTaq polymerase. To optimize the cycling conditions, we trialed 35, 38, 42, or 45 cycles, and annealing temperatures of 35˚C, 38˚C, 40˚C, 45˚C, or 48˚C.
3. Results and Discussion
In a preliminary study, we used DNA prepared from
Table 1. Plant materials used in this study.

three varieties of A. acutiloba (Toki, Hokkai Toki, and Miyama Toki) as templates, and screened 80 10-mer random primers (OPA, OPB, OPC, and OPD series) to investigate whether they detected polymorphisms. Of the 80 10-mers, 47 did not detect polymorphisms, 29 did not amplify all DNA templates isolated from the three varieties, and 4 gave highly reproducible polymorphic RAPD amplification patterns. From the four primers that de tected polymorphisms, we selected three for further analysis of plant materials as they produced the most distinct and reproducible polymorphisms.
Next, we optimized the PCR conditions for the RAPD analysis, since the reaction conditions can affect the pattern of PCR products [17]. The optimized conditions were as follows: 200 ng DNA, 2.5 U ExTaq polymerase, 42 cycles, and 40˚C annealing temperature (data not shown).
We obtained reproducible amplified DNA fragments for 14 varieties of A. acutiloba and 2 species of A.
Table 2. Nucleotide sequences of random primers used in this study.

stenoloba and A. shikokiana, using three primers (OPA- 16, OPD-3, and OPD-15). A. acutiloba varieties and A. stenoloba had many bands that differed from those of A. shikokiana (Figure 1). All of the A. acutiloba var. acutiloba strains showed the same RAPD profile using the OPD-3 primer, except for A. acutiloba var. acutiloba collected from Yamagata. The 1.6-kbp band observed in the fragment profile of A. acutiloba var. acutiloba, A. acutiloba var. iwatensis collected from Miyagi, and A. stenoloba after amplification with the OPA-16 primer was absent from the profiles of A. acutiloba var. sugiyamae, A. acutiloba var. iwatensis collected from Yamagata, and A. stenoloba var. lanceolata. The fragment pattern of A. acutiloba var. iwatensis collected from Miyagi differed from that of A. acutiloba var. iwatensis collected from Yamagata. Therefore, A. acutiloba var. iwatensis showed geographic variations in its genetic polymerphisms, similar to the findings of a previous study [8]. In addition, the profiles of A. acutiloba var. iwatensis collected from Miyagi were quite similar to that of A. acutiloba var. acutiloba after amplification with the OPA-16 primer. This implied that A. acutiloba var. acutiloba prescribed as a crude drug is closely related to a geographical strain of A. acutiloba var. iwatensis. Another A. acutiloba var. iwatensis collected from Yamagata showed generally the same fragment pattern as that of A. acutiloba var. sugiyamae after amplification with the OPD-15 primer, but the patterns differed after amplification with the OPA-16 primer. These results indicated that there was geographical variation among varieties of A. acutiloba var. iwatensis, and the genetic background of one of the varieties of A. acutiloba var. iwatensis was very similar to that of A. acutiloba var. sugiyamae. Among three strains of A. acutiloba var. sugiyamae, the slight differences in their fragment patterns after amplification with the OPD-3 primer might reflect indigenous polymorphisms that occurred independently during longterm maintenance of these traditional strains.
A. acutiloba var. acutiloba and A. acutiloba var. sugiyamae were discriminated by polymorphic bands generated by the OPA-16 primer. The fragment pattern of A. acutiloba var. sugiyamae after amplification with the OPD-15 primer contained a characteristic 1.1-kbp band. This was not found in A. acutiloba var. acutiloba strains, except for the strain collected from Yamagata. Therefore, this fragment could be useful to clarify whether the Japanese Angelica Root material is derived from A. acutiloba var. acutiloba or A. acutiloba var. sugiyamae. Although the same fragment was found in the Yamagata strain of A. acutiloba var. acutiloba using the OPD-15 primer, this strain showed a similar fragment pattern to that of other A. acutiloba var. acutiloba strains after amplification with the OPD-3 primer. Since previous studies have shown that the sequences of three regions in the chloroplast genome of A. acutiloba var. acutiloba from Yamagata were identical to those of A. acutiloba var. sugiyamae [8], these results demonstrated that the Yamagata strain was an interspecific hybrid derived from outcrossing between A. acutiloba var. acutiloba and A. acutiloba var. sugiyamae.
Authentic identification of A. acutiloba varieties is necessary for customers using these materials as a crude drug for traditional Japanese medicine to ensure the quality of the Japanese Angelica Root material. The traditional methods to authenticate “Toki”, “Hokkai Toki” and their adulterants are mainly based on morphological characters of the plants. In general, the differences in morphological characters and certain active compounds are subtle and ambiguous [18]. The RAPD analysis of A. acutiloba and related species represents an efficient method to authenticate Japanese Angelica Root, and confirm whether it is derived from “Toki” or “Hokkai Toki”. The molecular authentication of these regions is a highly sensitive and stable method. The authentication results were reliable and were not affected by the physical form or physiological conditions of the plant samples. Therefore, the RAPD marker method described in this study could be used for practical and accurate authentication of A. acutiloba var. acutiloba and A. acutiloba var. sugiya-
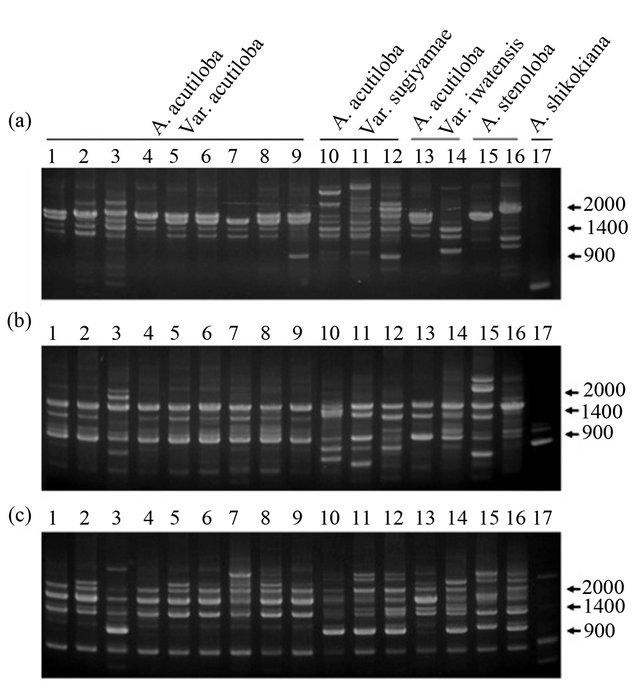
Figure 1. RAPD profiles generated by OPA-16 (a), OPD-3 (b) and OPD-15 (c) primers. Lanes 1 - 9: A. acutiloba var. acutiloba (TG, TK, TY, TH, TC, YB, YM, OT, OC; refer to Table 1 for codes), lanes 10 - 12: A. acutiloba var. sugiyamae (HA, HB, HC), lanes 13 and 14: A. acutiloba var. iwatensis (MM, MY), lane 15: A. stenoloba (LH), lane 16: A. stenoloba f. lanceolata (LT), lane 17: A. shikokiana (IM).
mae.
The Miyagi strain of A. acutiloba var. iwatensis showed a similar genetic background to A. acutiloba var. acutiloba and the Yamagata strain of A. acutiloba var. iwatensis showed a similar genetic background to A. acutiloba var. sugiyamae. These findings suggested that A. acutiloba var. iwatensis was a historical parent of both cultivars of A. acutiloba var. acutiloba and A. acutiloba var. sugiyamae. These RAPD analyses can be used to elucidate the origins of both A. acutiloba var. acutiloba and A. acutiloba var. sugiyamae. Since A. acutiloba var. iwatensis grows wild in Japan [6], further studies are required to clarify the history of outcrossing among these closely related species and varieties.
4. Acknowledgements
We are grateful to Prof. K. Koike, Faculty of Pharmaceutical Sciences, Toho University, Mr. H. Hayasaka, Graduate School of Pharmaceutical Science, Tohoku University, Mr. M. Murakami, Toyama Prefectural Institute for Pharmaceutical Research, and Dr. T. Shibata, Research Center for Medicinal Plant Resources, National Institute of Biomedical Innovation, for supplying A. acutiloba plants. We thank Assoc. Prof. K. Hotta, Faculty of Pharmaceutical Sciences, Health Sciences University of Hokkaido, for providing A. acutiloba var. sugiyamae plants.
NOTES